Studies on YibK and YbeA
Despite the vast topological diversity observed in biological structures, it was thought improbable, if not impossible,
that a polypeptide chain could 'knot' itself to form a functional protein. Nevertheless such knotted structures have been recently identified,
raising questions about how such complex topologies can arise during folding. Their formation does not fit any current folding models or
mechanisms and therefore represents an important piece of the protein-folding puzzle.
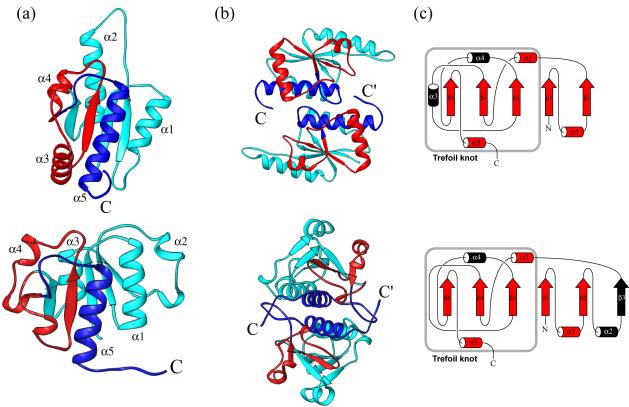
Experimental studies in our lab have primarily examined the folding of two of the smallest homodimeric α/β
MTases identified, YibK from Haemophilus influenzae and YbeA from Escherichia coli, both of which contain a trefoil knot in their backbone structure.
Using these model systems, we are exploring the mechanisms involved in protein knotting and folding.
Leading References
- Mallam AL, Onuoha SC, Grossmann JG, Jackson SE (2008) "Knotted Fusion Proteins Reveal Unexpected Possibilities in Protein Folding." Molecular Cell 30:642-648. PDF
- Mallam AL, Jackson SE (2007) "The dimerization of an α/β-knotted protein is essential for structure and function." Structure 15(1):111-122. PDF
- Mallam AL, Jackson SE (2007) "A comparison of the folding of two knotted proteins: YbeA and YibK." J. Mol. Biol. 366(2):650-665. PDF
- Mallam AL, Jackson SE (2006) "Probing nature's knots: the folding pathway of a knotted homodimeric protein." J. Mol. Biol. 359(5):1420-1436. PDF