Single molecule studies on the folding of ubiquitin
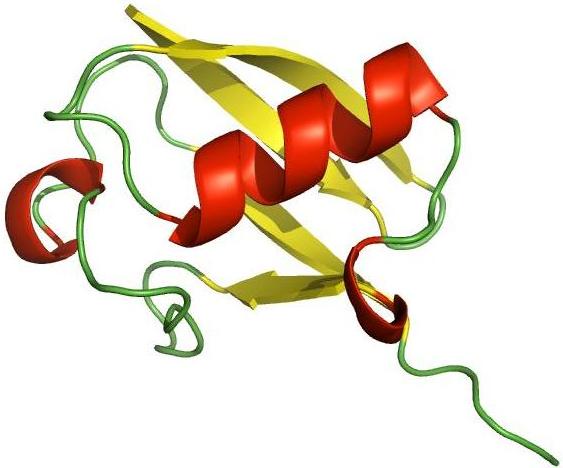
The small 76 residue, ubiquitous protein ubiquitin has been recognised as an essential part of life, taking a central role in controlled protein degradation by the 26S proteasome1. It is highly conserved from yeast to plants to mankind and is involved in vast numbers of cellular processes. We are exploring a variety of labelling strategies and fluorophores for optimal dual-labelling of ubiquitin, a key issue in application of single molecule fluorescence methods to protein folding/unfolding. Using this dual-labelled sample we are studying the unfolding and refolding of ubiquitin at the single molecule level employing a specially designed single molecule nanomixer2. Recently, we extended this methodology to monitor single-molecule refolding, by adding a second micropipette to the nanomixer for double-jump experiments. These experiments are likely to shed some light on the question whether ubiquitin is a classical two-state folder or if there is a more complex energy landscape with intermediate(s) on or off the folding pathway.
Leading References
- Hershko A, Ciechanover A, Rose IA. (1979) "Resolution of the ATP-dependent proteolytic system from reticulocytes: A component that interacts with ATP". Proc. Natl. Acad. Sci. USA, 76:3107-3110
- Orte A, Craggs TD, White SS, Jackson SE, Klenerman D. (2008) "Evidence of an intermediate and parallel pathways in protein unfolding from single-molecule fluorescence". J. Am. Chem. Soc., 130(25):7898-7907
- Jackson SE (2008) "The Solution to Multiple Structures." Structure 16:659-661. PDF